In this post we describe how to do regression with count data using R.
In many applications we want to model the relationship between features and responses where the responses take count values. For instance, we may want to model the number of times an event happens in a fixed interval, given covariates. We might want to model the number of cigarettes smoked, given context, the number of calls arriving at a call center as a function of business characteristics, or the number of people arriving at a location, given characteristics of a location.
A first thought is to use linear regression naively: this poses problems including skewness, use of a continuous distribution, and interpretation. A second idea is to use a Poisson distribution to model ![]() , where
, where ![]() is the design matrix of features. However, the Poisson distribution has the limitation that its variance equals its mean: fitting it often leads to overdispersion, where the observed variance is higher than that expected by the model. One can handle this in two ways: by quasi-likelihood estimation, or by modeling
is the design matrix of features. However, the Poisson distribution has the limitation that its variance equals its mean: fitting it often leads to overdispersion, where the observed variance is higher than that expected by the model. One can handle this in two ways: by quasi-likelihood estimation, or by modeling ![]() with a negative binomial distribution, which generalizes the Poisson distribution to two parameters, allowing the variance to differ from the mean. A final common problem is zero inflation, where the observed number of
with a negative binomial distribution, which generalizes the Poisson distribution to two parameters, allowing the variance to differ from the mean. A final common problem is zero inflation, where the observed number of ![]() ‘s is higher than that expected by either a Poisson or negative binomial distribution: this can be solved via zero inflated models. In this article, we describe each modeling approach, its limitations, how we can handle those limitations, and show examples in R.
‘s is higher than that expected by either a Poisson or negative binomial distribution: this can be solved via zero inflated models. In this article, we describe each modeling approach, its limitations, how we can handle those limitations, and show examples in R.
Using Linear Regression
Without Transforming Response
We can naively fit a linear regression model
(1) ![]()
here ![]() is the vector of count data observations and
is the vector of count data observations and ![]() is a design matrix of features. The first problem with this formulation is that our true
is a design matrix of features. The first problem with this formulation is that our true ![]() are lower bounded by
are lower bounded by ![]() , while our fitted values
, while our fitted values ![]() are not. Thus we will have poor predictive accuracy and inferences about behavior close to
are not. Thus we will have poor predictive accuracy and inferences about behavior close to ![]() . Further, when it comes to hypothesis testing, unless we have a lot of data, the skewness of
. Further, when it comes to hypothesis testing, unless we have a lot of data, the skewness of ![]() will be problematic. This is generally a bad model, although it can be less bad in cases where all the observed
will be problematic. This is generally a bad model, although it can be less bad in cases where all the observed ![]() are far from
are far from ![]() and the true
and the true ![]() has low skewness.
has low skewness.
Taking Log of Response
We can also fit
(2) ![]()
Under this setting for hypothesis testing we assume (and need to test) that ![]() is log-normal. This is good in that it captures the skewness often present in count data, and
is log-normal. This is good in that it captures the skewness often present in count data, and ![]() is also is now lower bounded by
is also is now lower bounded by ![]() . However, this model also presents a few issues. Firstly, one fits a continuous distribution to discrete data, leading to questionable inference results.
. However, this model also presents a few issues. Firstly, one fits a continuous distribution to discrete data, leading to questionable inference results.
Let’s look at the interpretation in the best case, where ![]() never has any
never has any ![]() values and thus we can fit
values and thus we can fit ![]() . In this case a unit change in
. In this case a unit change in ![]() is associated with an
is associated with an ![]() change in
change in ![]() , holding noise fixed, i.e. changes
, holding noise fixed, i.e. changes ![]() by
by ![]() . This has two problems: 1) we don’t want
. This has two problems: 1) we don’t want ![]() to take arbitrary real values and 2) the holding noise fixed interpretation is not intuitive at all.
to take arbitrary real values and 2) the holding noise fixed interpretation is not intuitive at all.
Poisson Regression
An alternative to using linear regression is to use Poisson regression, which assumes that ![]() . This is a generalized linear model, and the equation to represent this is
. This is a generalized linear model, and the equation to represent this is
(3) ![]()
The interpretation becomes a unit change in ![]() is associated with an
is associated with an ![]() multiplicative change in
multiplicative change in ![]() . This treats discrete observations with a discrete distribution and is more intuitive than log-transforming the response.
. This treats discrete observations with a discrete distribution and is more intuitive than log-transforming the response.
Let’s fit a Poisson regression model to the warpbreaks dataset in R
pois_reg <- glm(formula = breaks ~ wool+tension, data = warpbreaks,
family = poisson)
summary(pois_reg)
Call:
glm(formula = breaks ~ wool + tension, family = poisson, data = warpbreaks)
Deviance Residuals:
Min 1Q Median 3Q Max
-3.6871 -1.6503 -0.4269 1.1902 4.2616
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 3.69196 0.04541 81.302 < 2e-16 ***
woolB -0.20599 0.05157 -3.994 6.49e-05 ***
tensionM -0.32132 0.06027 -5.332 9.73e-08 ***
tensionH -0.51849 0.06396 -8.107 5.21e-16 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
(Dispersion parameter for poisson family taken to be 1)
Null deviance: 297.37 on 53 degrees of freedom
Residual deviance: 210.39 on 50 degrees of freedom
AIC: 493.06
Number of Fisher Scoring iterations: 4
Zero-Inflation
In practical datasets, one often sees more zeros than would be predicted by a Poisson distribution. For instance, say our ![]() is the number of cigarettes smoked in the first day of a quit attempt for some participant
is the number of cigarettes smoked in the first day of a quit attempt for some participant ![]() . Often this participant will come from one of two groups: those who successfully quit and those who don’t. If those who don’t have a Poisson distribution as their true distribution for number of cigarettes smoked, then the distribution across all participants will have more
. Often this participant will come from one of two groups: those who successfully quit and those who don’t. If those who don’t have a Poisson distribution as their true distribution for number of cigarettes smoked, then the distribution across all participants will have more ![]() s than posited by a Poisson.
s than posited by a Poisson.
We can model this via a mixture model with two components, where
(4) 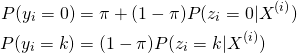
here ![]() is the probability of being in component
is the probability of being in component ![]() , which takes value
, which takes value ![]() with probability
with probability ![]() , and
, and ![]() is the probability of being in component
is the probability of being in component ![]() , and
, and ![]() is a Poisson distributed random variable with mean
is a Poisson distributed random variable with mean ![]() . In order to test whether this is a good model, we can use the hypothesis test
. In order to test whether this is a good model, we can use the hypothesis test
 , that is, the component that takes
, that is, the component that takes  with probability
with probability  has
has  weight
weight , this component has non-zero weight.
, this component has non-zero weight.
We can use a score test here [1], which can be found in the R package countreg. Let’s perform this test on the warpbreaks dataset in R (which incidentally doesn’t have any ![]() values in the responses).
values in the responses).
library(countreg)
zitest(pois_reg)
Zero inflation test
data: pois_reg
S = 1.2158e-15, p-value = 0.5
As expected, we do not have a significant p-value at any of the standard cutoffs.
Overdispersion
Another issue often found in practice is that real data has overdispersion, where the variance is higher than the mean, while the Poisson distribution has variance equal to the mean. A common technique to ‘detect’ this is via a deviance goodness of fit test. However, this only tells us how good a fit a Poisson model is, not whether overdispersion is present.
An alternative is to instead use negative binomial regression. The negative binomial distribution has an additional parameter, allowing both the mean and variance to be estimated. Since the Poisson distribution is a special case of the negative binomial and the latter has one additional parameter, we can do a ![]() test where
test where
 : true model is Poisson regression
: true model is Poisson regression : true model is negative binomial regression
: true model is negative binomial regression
Let’s test for overdispersion in R on the warpbreaks dataset.
library(pscl)
nb_reg <- glm.nb(formula=breaks ~ wool+tension, data = warpbreaks)
odTest(nb_reg)
Likelihood ratio test of H0: Poisson, as restricted NB model:
n.b., the distribution of the test-statistic under H0 is non-standard
e.g., see help(odTest) for details/references
Critical value of test statistic at the alpha= 0.05 level: 2.7055
Chi-Square Test Statistic = 86.2922 p-value = < 2.2e-16
And thus we reject the null hypothesis that the true model is Poisson regression in favor of negative binomial regression.
Putting it Together
If you didn’t notice, we performed two hypothesis tests here: one for a zero inflated model, and one for a negative binomial model. We thus need to adjust our p-value threshold. Instead of using some value ![]() , we have to do
, we have to do ![]() , where
, where ![]() is the number of hypothesis tests we ran. This is the Bonferroni correction.
is the number of hypothesis tests we ran. This is the Bonferroni correction.
From our tests, we decided to use a negative binomial model without using a zero-inflated model. Let’s look at the results.
summary(nb_reg)
Call:
glm.nb(formula = breaks ~ wool + tension, data = warpbreaks,
init.theta = 9.944385436, link = log)
Deviance Residuals:
Min 1Q Median 3Q Max
-2.0144 -0.9319 -0.2240 0.5828 1.8220
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 3.6734 0.0979 37.520 < 2e-16 ***
woolB -0.1862 0.1010 -1.844 0.0651 .
tensionM -0.2992 0.1217 -2.458 0.0140 *
tensionH -0.5114 0.1237 -4.133 3.58e-05 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
(Dispersion parameter for Negative Binomial(9.9444) family taken to be 1)
Null deviance: 75.464 on 53 degrees of freedom
Residual deviance: 53.723 on 50 degrees of freedom
AIC: 408.76
Number of Fisher Scoring iterations: 1
Theta: 9.94
Std. Err.: 2.56
2 x log-likelihood: -398.764
Note that if we consider p-values for each individual coefficient, we actually need a Bonferroni correction. We performed two hypothesis tests to arrive at our final model, and have three features and one intercept, leading to six hypothesis tests. In order to ensure the probability of no type I errors is less than ![]() , we look for individual p-values of
, we look for individual p-values of ![]() . Under this criteria we can consider the intercept and tensionH to be significant, but not the other features.
. Under this criteria we can consider the intercept and tensionH to be significant, but not the other features.
[1] Van den Broek, Jan. “A score test for zero inflation in a Poisson distribution.” Biometrics (1995): 738-743.