
In survival analysis, we want to model the time to a first event, often death. One way to model the risk is using Cox regression, which we describe in this post. We then show a simple example.
We describe the instantaneous risk of an event via a function called the hazard. We would like to describe how this risk changes over time. We’re interested in risk changes at a global level across patients, and at an individual level in terms of covariates. Covariates can be weight, age at the start of the study, what treatment they receive, etc. Cox regression [1], a technique for describing this, specifies a global baseline hazard, which is non-parametric and thus allows the global risk to vary in a very flexible way as a function of time, and a relative risk that describes how covariates influence risk and is parametric. However, this semi-parametric specification makes inference difficult: in this article we describe this model and David Cox’s insight for how to do inference in it.
Basics of Survival Analysis
In survival analysis we want to model the probability of survival until at least time ![]() . Let
. Let ![]() be the death time. Then we want to model
be the death time. Then we want to model
(1) ![]()

where ![]() is the cumulative distribution function of the death time
is the cumulative distribution function of the death time ![]() . The hazard is an important concept here. It is the instantaneous risk of death at time
. The hazard is an important concept here. It is the instantaneous risk of death at time ![]() , conditional on surviving up to
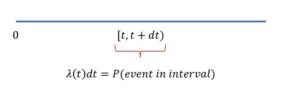
, conditional on surviving up to ![]() . For a heuristic derivation, let the conditional probability of death in the small interval
. For a heuristic derivation, let the conditional probability of death in the small interval ![]() be given by
be given by ![]() . By rearranging terms and taking limits we obtain
. By rearranging terms and taking limits we obtain
(2) ![]()
with some manipulation we can relate these two for continuous distributions via
(3) ![]()
The Cox Hazard and its Likelihood
In order to compute this, we need to specify a functional form for the hazard. Cox proposed that for an individual ![]() ,
,
(4) ![]()
where ![]() , the baseline risk, is often taken to be non-parametric and
, the baseline risk, is often taken to be non-parametric and ![]() is a vector of covariates for the individual
is a vector of covariates for the individual ![]() . As mentioned in the introduction, the non-parametric baseline risk allows the global risk shared across patients to vary in a very flexible way, while the parametric relative risk allows us to incorporate individual covariates. However, this specification prevents us from doing maximum likelihood estimation directly. To see why, let
. As mentioned in the introduction, the non-parametric baseline risk allows the global risk shared across patients to vary in a very flexible way, while the parametric relative risk allows us to incorporate individual covariates. However, this specification prevents us from doing maximum likelihood estimation directly. To see why, let ![]() be
be ![]() if the patient has their death observed and
if the patient has their death observed and ![]() otherwise (for an example of the latter, if they survive until the end of the study or drop out). Then letting
otherwise (for an example of the latter, if they survive until the end of the study or drop out). Then letting ![]() be the density for participant
be the density for participant ![]() and
and ![]() be their survival function, the likelihood is the product of the densities for observed death times and the survival probabilities for censored patients. This gives
be their survival function, the likelihood is the product of the densities for observed death times and the survival probabilities for censored patients. This gives
(5) ![Rendered by QuickLaTeX.com \begin{align*}\prod_{n=1}^N f_n(t)^{\delta_n}S_n(t)^{1-\delta_n}&=\prod_{n=1}^N (-S_n'(t))^{\delta_n} S_n(t)^{1-\delta_n}\\&=\prod_{n=1}^N [(\lambda_n(t) S_n(t)]^{\delta_n} S_n(t)^{1-\delta_n}\\&=\prod_{n=1}^N [\lambda_0(t)\exp(\beta^T z_n)]^{\delta_n} S_n(t)\end{align*}](https://boostedml.com/wp-content/ql-cache/quicklatex.com-2a37c04cea41ac083c21f94052329cd8_l3.png)
The Partial Likelihood
Note that in the previous likelihood, we have the nuisance parameter ![]() , which is non-parametric. We thus cannot directly maximize the entire likelihood directly to estimate the parameters
, which is non-parametric. We thus cannot directly maximize the entire likelihood directly to estimate the parameters ![]() . However, we can express the above likelihood as follows
. However, we can express the above likelihood as follows
(6) 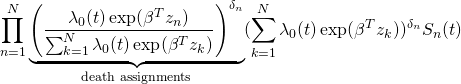
Cox’s insight was that the first term, the death assignment probabilities, given event times, contains most of the information about ![]() , while the remaining term contains most of the information about
, while the remaining term contains most of the information about ![]() . We can thus maximize the product of the first terms, the partial likelihood: the part of the likelihood that contains most of the information about
. We can thus maximize the product of the first terms, the partial likelihood: the part of the likelihood that contains most of the information about ![]() . Later authors [2,3] proved that doing so gives consistent and asymptotically normal estimates
. Later authors [2,3] proved that doing so gives consistent and asymptotically normal estimates ![]() of
of ![]() .
.
An Example
We now show an example on the ovarian dataset from the survival package. First let’s have a look at the data
library(survival)
data(ovarian)
head(ovarian)
futime fustat age resid.ds rx ecog.ps
1 59 1 72.3315 2 1 1
2 115 1 74.4932 2 1 1
3 156 1 66.4658 2 1 2
4 421 0 53.3644 2 2 1
5 431 1 50.3397 2 1 1
6 448 0 56.4301 1 1 2
We see the variables
- futime: last observed time
- fustat: censoring indicator
- rx: presence of residual disease
- patient age
- rx: treatment group assignment
- ecog.ps: performance status, patient’s level of functioning in life
Let’s do Cox regression now, regressing death risk on age and performance status as our covariates, and look at the summary
fit <- coxph(Surv(futime, fustat) ~ age + ecog.ps, data=ovarian)
summary(fit)
Call:
coxph(formula = Surv(futime, fustat) ~ age + ecog.ps, data = ovarian)
n= 26, number of events= 12
coef exp(coef) se(coef) z Pr(>|z|)
age 0.16150 1.17527 0.04992 3.235 0.00122 **
ecog.ps 0.01866 1.01884 0.59908 0.031 0.97515
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
exp(coef) exp(-coef) lower .95 upper .95
age 1.175 0.8509 1.0657 1.296
ecog.ps 1.019 0.9815 0.3149 3.296
Concordance= 0.784 (se = 0.085 )
Likelihood ratio test= 14.29 on 2 df, p=8e-04
Wald test = 10.54 on 2 df, p=0.005
Score (logrank) test = 12.26 on 2 df, p=0.002
Only age has a significant coefficient. The interpretation is that holding performance status fixed, an increase in age of one year has a 1.18 times multiplicative effect on death risk. This intuitively makes sense. The next section gives confidence intervals for the multiplicative effects of the covariates. After that, the concordance tells us the probability that the survival prediction goes in the same direction as the actual data. That is, for two randomly observations, the probability that an observation with shorter survival time has higher risk score. This is a goodness-of-fit statistic.
The remaining tests have as ![]() : all coefficients are
: all coefficients are ![]() . As you can see, we reject this.
. As you can see, we reject this.
[1] Cox, David R. “Regression models and life-tables.” In Breakthroughs in statistics, pp. 527-541. Springer, New York, NY, 1992.
[2] Tsiatis, Anastasios A. “A large sample study of Cox’s regression model.” The Annals of Statistics 9, no. 1 (1981): 93-108.
[3] Andersen, Per Kragh, and Richard David Gill. “Cox’s regression model for counting processes: a large sample study.” The annals of statistics (1982): 1100-1120.