In this post we describe how to solve the full rank least squares problem without inverting a matrix, as inverting a matrix is subject to numerical stability issues. We first describe the least squares problem and the normal equations, then describe the naive solution involving matrix inversion and describe its problems. We then describe two other methods: the Cholesky decomposition and the QR decomposition using householder matrices. The first is also unstable, while the second is far more stable. We show examples in python, using numpy and scipy.
The Least Squares Problem
Let ![]() , and let
, and let ![]() be full rank. The least squares problem is to solve
be full rank. The least squares problem is to solve
(1) ![]()
the main application of this is in linear regression, although in that setting it’s generally written as
(2) ![]()
we will use the former notation, which is common in the numerical linear algebra literature.
Normal Equations
In order to solve this, we can expand the terms in the norm and take derivatives, giving us the normal equations:
(3) ![]()
the simplest way to solve this is to compute ![]() and then compute
and then compute
(4) ![]()
however let ![]() be the singular values of
be the singular values of ![]() . If
. If ![]() is close to
is close to ![]() , then the computation of
, then the computation of ![]() will have large errors. Let’s try an extreme case where the matrix
will have large errors. Let’s try an extreme case where the matrix ![]() is almost singular, and the number of rows is equal to the number of columns.
is almost singular, and the number of rows is equal to the number of columns.
import numpy as np
from scipy import linalg
np.random.seed(3)
m = 1000
n = 1000
Cov = 0.999999*np.ones((n,n))
np.fill_diagonal(Cov,1)
A = np.random.multivariate_normal(np.zeros(n),Cov,m)
b = np.random.normal(0,1,m)
gram = np.dot(A.T,A)
print(np.linalg.cond(gram))
6.392675702631713e+17
this has extremely high condition number, suggesting that many procedures, in particular matrix inversion, will be numerically unstable. Let’s try solving the least squares problem and computing the residual norm ![]() for our estimate
for our estimate ![]() .
.
x_normal_equations = np.dot(np.linalg.inv(gram),np.dot(A.T,b))
r_norm = np.linalg.norm(np.dot(gram,x_normal_equations)-np.dot(A.T,b))
print(r_norm)
35.13589871545292
this is a huge error! Clearly we can’t use this method as an all purpose solution.
Cholesky Decomposition
One alternative to inverting the matrix is to do a Cholesky decomposition ![]() , where
, where ![]() is lower triangular. We can then solve
is lower triangular. We can then solve
(5) ![]()
we can then solve the triangular system ![]() for
for ![]() and
and ![]() for
for ![]() . Let’s see how this does.
. Let’s see how this does.
gram = np.matrix(gram)
L = np.linalg.cholesky(gram)
y = linalg.solve_triangular(L,A.T.dot(b),lower=True)
x_cholesky = linalg.solve_triangular(L.T,y)
r_norm_cholesky = np.linalg.norm(np.dot(gram,x_cholesky)-np.dot(A.T,b))
print(r_norm)
35.13589871545292
this is the same error. Clearly we can’t use this either.
Householder QR
Householder QR finds a sequence of householder matrices ![]() such that
such that ![]() , where
, where ![]() is upper triangular, and
is upper triangular, and ![]() , where
, where ![]() is an orthogonal matrix. Thus
is an orthogonal matrix. Thus ![]() is a
is a ![]() decomposition. This is more stable than a QR decomposition using classical gram schmidt.
decomposition. This is more stable than a QR decomposition using classical gram schmidt.
We can plug this into the normal equations via
(6) 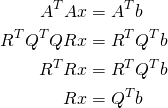
we can then solve the triangular system ![]() for
for ![]() .
.
Q,R=np.linalg.qr(A)
x_householder=linalg.solve_triangular(R,Q.T.dot(b))
r_norm_householder = np.linalg.norm(np.dot(gram,x_householder)-np.dot(A.T,b))
print(r_norm_householder)
5.681399611646504e-05
This is far more stable!
Discussion
In this post we describe three methods for solving the normal equations for full rank ![]() . The first two are particularly vulnerable to numerical errors, while the third is more robust.
. The first two are particularly vulnerable to numerical errors, while the third is more robust.